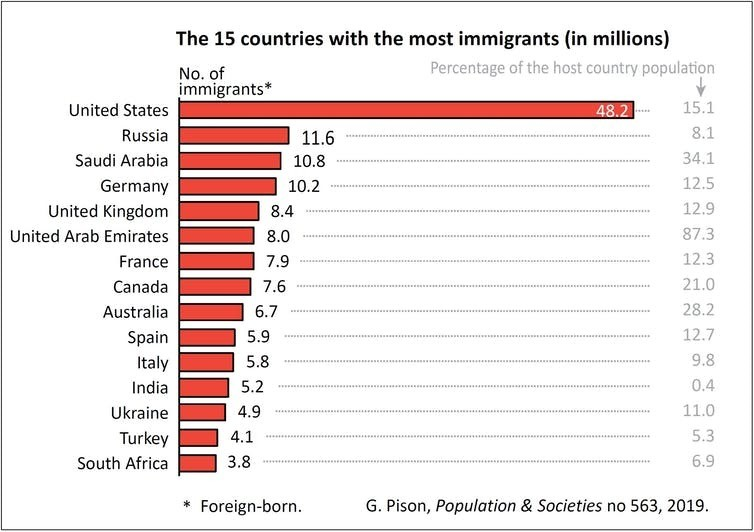
The Technical Infrastructure team at Google Cloud recently came to our school with a question about the scale of its biggest competitor, Amazon Web Services (AWS).
To summarize, Amazon recently swapped out the depreciation schedule of its servers from 3 years to 4 years, straight line. This is a big deal since extending a depreciation schedule means significantly less reported expenses each quarter while that change takes effect. Specifically, they reported a difference of $800 million for the next quarter alone.
It turns out you could actually work out a lot about Amazon’s server spending by just crunching some numbers over the past 7 years. In short, you could estimate the portion of capital expenditures allocated to servers by calculating what spending schedule would sum to the $800 million figure (assuming that the depreciation change was implemented) and work backwards. Sounds complicated, but it’s easier than you think.
So here’s the $800 million accounting question: From that figure and other publicly available data (SEC filings, earnings statements, industry reports, etc.), would it be possible to reverse engineer how much Amazon spends on their servers, and thus, get an idea of how many they currently have in their fleet?
This problem was the impetus for our school hosting a 6-hour hackathon with Google to see who could come up with the best answer. We eventually took home first prize (go team Lime Green)! Here’s what we did.
Theory & Background
Why does Google want this info? A good estimate of AWS’s scale could help a competitor understand where they stand in terms of theoretical capacity (not just market share), how they compare in spending, and make predictions of an approximate rate of expansion through a time series.
In turn, AWS is fairly secretive about exactly how many servers they have and where their data centers are even located. I mean, why wouldn’t they be?
Despite this, we can derive a pretty good estimate using figures from their SEC reports and, crucially, that figure they released in their most recent earnings call (the $800 million effect). If we set that as a constant, we can do some basic Excel math to work backward and approximate each quarter’s spending on servers. From there, we can estimate the server fleet size by dividing that spending by approximate cost-per-server adjusted by year (server costs have changed a lot since 2014).
It’s far from perfect, but it’s not a bad starting point for uncovering market intelligence intended to be kept hidden using data that’s widely available. Here’s the idea:
- Using past 10-Q reports, scrape Amazon’s capital expenditures (CapEx) for the past 3 years/12 quarters (those affected by the accounting change). The idea here is that server spending will fall under CapEx, but the breadth of that category usually “hides” it with other expenses.
- Calculate how much each quarter’s depreciation expense would be affected by the change in schedule. e.g. if it was purchased just last quarter, it would change from the total 3 to 4 years remaining or 12 to 16 quarters. If two quarters ago, then it would change from 11 to 15 quarters, and so on.
- Make an assumption about relative spending. There’s too many unknowns since we don’t know what portion was spent on servers within each quarter. The simplest is to assume that the percent CapEx allocated to servers is constant. There are other possibilities, though adjusting this parameter doesn’t turn out to be that important compared to the other assumptions.
- Finally, determine a moving estimate of the average server cost for a firm like Amazon based on various assumptions (how long servers last, OEM server manufacturer averages, how long it takes to expense them, potential fixed costs, where/how AWS servers are manufactured, etc) and market trends adjusted for each year. Divide and get an estimate for server fleet size. Done!
Writeup
Okay, let’s see it in action. What follows is a slightly edited version of the deliverable that won us the hackathon. If you want to follow the modeling aspect, you’ll also need this excel sheet (596 downloads ) .
We only had six hours to write in the actual competition, but we had the chance to refine some things afterward in preparation for a presentation with the Google Cloud team. Thus, there’s also extra considerations like subtracting the cost of server racks and adjusting for DRAM prices (the main cost-driver of servers):
Estimating the Scale of Amazon’s Server Operation
A Market Intelligence Report based on SEC Filings, Earnings Statements, Industry Reports, and Other Public Information
Submitted for Consideration in the 2020 MQM Forensics Hackathon
Team Lime Green – Shay Xueyao Fu, Shangyun Song, Yingli Sun, and Alex Zhou
What We Know
Amazon recently extended the depreciation schedule of its servers from a straight-line 3 years schedule to 4 years, which will result in an $800 million decreased depreciation expense. Amazon’s total quarterly capital expenditures are made publicly available from SEC filings. AWS currently accounts for about 71% of Amazon’s operating profit.
IDC-published results put ODM servers at an average cost of $6,486.55 each for Q3 of 2019, which are about in line with recent averages for x86 server selling prices. AWS uses custom-made white box systems (likely designed in-house and produced by an ODM) and is even building a repertoire of custom silicon for servers, likely driving unit cost down. From the same published results, we can obtain the total revenue and market share of key players and ODM direct sources in the server market for each quarter.
What We’ll Assume
Since we cannot isolate the relative CapEx set aside for servers in each quarter by Amazon based on SEC filings, we assumed two possible spending schedules: a constant-rate schedule and a market-adjusted schedule. The constant schedule makes the simplest assumption that Amazon’s server spending does not change much year-over-year.
The market-adjusted schedule uses trends in ODM server revenue by quarter and adjusts Amazon’s predicted spending based on this growth, as well as considering the growth rate of AWS availability zones and the change in DRAM pricing. Additionally, we subtract the cost of racks and network switches from the CapEx in both schedules when we calculate the number of servers.
While this assumption is not perfect and Amazon’s spending could differ from market trends, this helps to account for explosive growth in the ODM server market (which is driven not in small part by AWS). The ODM market’s expansion, combined with nonlinear revenue growth for AWS in recent years, gives us reason to challenge a constant percent assumption for Amazon’s server spending. We provide estimates based on both assumptions in our appendix.
What We Found
Using the $800 million decrease, we estimate Amazon’s percent CapEx spent on servers to be 53.03% based on the constant-rate schedule and 47.34% from the market-adjusted schedule. Over the past 6 years, we estimate Amazon’s server spending to total $28.23 billion according to the constant-rate and $25.20 billion with the market-adjusted rate. Adjusting for average ODM server prices starting from 2014 and assuming an average useful life of 7 years and using both a floating/constant server price, Amazon currently has approximately 4.9 to 5.5 million servers in operation. This is in line with another estimate we produced based on the approximate amount of servers per data center per availability zone.
Appendix
Percent of Amazon’s (and/or AWS) CapEx spent on servers
We created two excel models and used Solver to find the base percent of Amazon’s CapEx going towards servers. CapEx was set as the “purchases of property and equipment, including internal-use software and website development, net” from each quarterly and annual filings’ consolidated statements of cash flows.
The first model was based on the Amazon quarterly CapEx spending which can be found from Amazon websites under investor relation. The second model considered the market adjusted schedule taken from the total revenue of the ODM server market from IDC, as well as considering the growth rate of AWS availability zones and the change in DRAM pricing.
Reference used:
Fixed percent of CapEx on servers:
Market-adjusted floating percent of CapEx on Servers:
How Much Amazon has Spent on Servers over the Last 6 years
We used the base rate calculated from question 1 multiplied by the yearly CapEx spending found from Amazon 10k.
Reference used:
- Amazon SEC filings: https://ir.aboutamazon.com/sec-filings
Number of Servers AWS Currently Has in Their Fleet
We must assume servers last between 5-8 years (competition guidelines). We’ll pick the higher end of this scale due to Amazon’s lengthy tenure in the server market and simplify calculations by choosing a constant server lifetime of 7 years. This means all servers purchased from 2014 (and no earlier) should still be in operation today.
We used average ODM server prices from each year starting from 2014 to estimate the cost Amazon paid to their manufacturers. We also considered adjusting the CapEx spending for network switches and racks to calculate the number of servers.
Another way to estimate server count is to use the number of server availability zones (69) and then approximate the number of datacenters per availability zone and the number of servers per datacenter. Estimates given at a conference by AWS engineer James Hamilton place the range of servers per data center at 50,000-80,000, and the number of centers per availability zone somewhere between 1-6. We still need to consider the cost of racks and network switches that are recorded as servers. From these ranges and educated guesses from the article, we can determine a current server count using the number of availability zones.
69[1 to 6 DCs/AZ][50000 to 65000 Servers/DC] = 3.4 million to 26.9 million servers
This upper bound of the base estimate is likely high since newly established AZs should have fewer average data centers than the ones referenced when the article was published. Each data center should also have fewer than the maximum number of servers as they ramp up. Likewise, our final estimate based on the financials resulted in 4.9 to 5.5 million servers, depending on whether or not we adjusted for ODM server prices in the market and other uncertain factors.
The calculation can be found at the screenshot attached to question 2 and our Excel workbook.
References Used:
- ODM server price in 2014- 2015
- For server price in the years 2016, we took the middle number from the range $2805-$6500, which is ~ $4500 in our calculation.
- ODM server price in 2017 and 2018
- ODM server price in 2019
- CapEx spending on server adjusted by spending on network and racks
- Cost of racks and networks
- DRAM pricing
- Amazon availability zones
- Availability zone launching announcement